Dr. Damien F. Meyer

Microbial Genomics Scientist & UX Researcher (PhD, HDR)
CIRAD, Guadeloupe
📧 damien.meyer@cirad.fr
“If we are to understand dynamics of bacterial-host cell interactions, we need to investigate the intimate adaptations at the cellular and molecular levels”
About Me
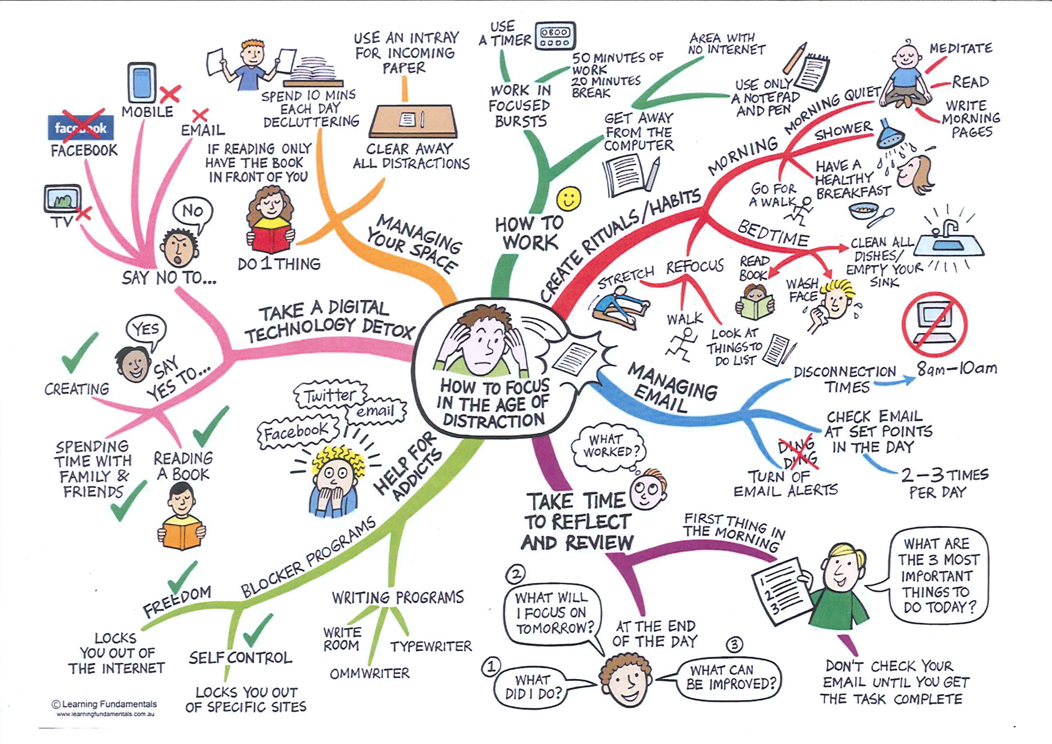
I am a senior researcher at CIRAD specializing in bacterial genomics and UX research applied to molecular microbiology. My work focuses on host-pathogen interactions, genome evolution, and data visualization.
I am also passionate about scientific communication and the application of design thinking in computational biology.
My research encompasses the molecular mechanisms that enable bacterial pathogens to modulate host cell functions and evade immune responses. To this end, I apply a systems biology approach that combines the power of bioinformatics with the precision of molecular and cellular biology to understand how pathogenic bacteria manipulate host cell signaling pathways.

All scientists are welcome to apply for a position in the lab, regardless of background or training experience in bacteriology or entomology. We seek proactive and collaborative lab members, particularly those with computational biology skills, to contribute to our interdisciplinary research.
Research Interests
- Gene evolution in pathogenic bacteria
- Secretion systems and bacterial virulence
- Bacterial phylodynamics
- Predictive bioinformatics and visual tools for microbiologists
Professional Links
- ORCID: https://orcid.org/0000-0003-2735-176X
- Google Scholar: https://scholar.google.com/citations?user=6kZSwKEAAAAJ
- LinkedIn: https://www.linkedin.com/in/damien-meyer-phd-hdr-849831b/
- Portfolio (PDF): MEYER_portfolio_2024.pdf
Selected Publications
- Cabello, A.-L. et al. Brucella driven host N-glycome remodeling controls infection. Cell Host Microbe 32 (2024). https://doi.org/10.1016/j.chom.2024.03.003
- Meyer, D.F., Moumène, A. Microbe Profile: Ehrlichia ruminantium – stealthy as it goes. Microbiology 169, 001415 (2023). https://doi.org/10.1099/mic.0.001415
- Gordon, J.L. et al. Possible biased virulence attenuation in the Senegal strain of Ehrlichia ruminantium by ntrX gene conversion. PLoS One 18, e0266234 (2023). https://doi.org/10.1371/journal.pone.0266234
- Dereeper, A. et al. PanExplorer: a web-based tool for exploratory analysis and visualization of bacterial pan-genomes. Bioinformatics 38, btac504 (2022). https://doi.org/10.1093/bioinformatics/btac504
- Noroy, C., Meyer, D.F. The super repertoire of type IV effectors in the pangenome of Ehrlichia spp. provides insights into host specificity and pathogenesis. PLoS Comput. Biol. 17, e1008788 (2021). https://doi.org/10.1371/journal.pcbi.1008788
- Moumène, A. et al. Iron starvation upregulates Ehrlichia ruminantium type IV secretion system and map1 genes. Front. Cell. Infect. Microbiol. 7, 535 (2018). https://doi.org/10.3389/fcimb.2017.00535
- Tago, D., Meyer, D.F. Economic game theory to model the attenuation of virulence of an obligate intracellular bacterium. Front. Cell. Infect. Microbiol. 6, 86 (2016). http://dx.doi.org/10.3389/fcimb.2016.00086
- Meyer, D.F. et al. Searching algorithm for type IV secretion system effectors 1.0. Nucleic Acids Res. 41, 9218–9229 (2013). https://doi.org/10.1093/nar/gkt718
- Blanvillain, S. et al. Plant carbohydrate scavenging through TonB-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS One 2, e224 (2007). https://doi.org/10.1371/journal.pone.0000224
© 2025 Damien F. Meyer. All rights reserved.