My research
Gottin C, Dievart A, Summo M, Droc G, Périn C, Ranwez V, Chantret N. (2021) A new comprehensive annotation of leucine-rich repeat-containing receptors in rice. Plant J. 108(2):492-508. doi: 10.1111/tpj.15456
Dievart A*, Gottin C*, Périn C, Ranwez V, Chantret N. (2020) Origin and Diversity of Plant Receptor-Like Kinases. Annual Review of Plant Biology. 71:131-156
Bureau C, Lanau N, Ingouff M, Hassan B, Meunier AC, Divol F, Sevilla R, Mieulet D, Dievart A, Périn C. (2018) A protocol combining multiphoton microscopy and propidium iodide for deep 3D root meristem imaging in rice: application for the screening and identification of tissue-specific enhancer trap lines. Plant Methods. 14:96.
Bettembourg M, Dal-Soglio M, Bureau C, Vernet A, Dardou A, Portefaix M, Bes M, Meynard D, Mieulet D, Cayrol B, Perin C, Courtois B, Ma JF, Dievart A (2017) Root cone angle is enlarged in docs1 LRR-RLK mutants in rice. Rice 10:50.
The DEFECTIVE IN OUTER CELL LAYER SPECIFICATION 1 (DOCS1) gene belongs to the Leucine-Rich Repeat Receptor-Like Kinase (LRR-RLK) subfamily. It has been discovered few years ago in Oryza sativa (rice) in a screen to isolate mutants with defects in sensitivity to aluminum. The c68 (docs1-1) mutant possessed a nonsense mutation in the C-terminal part of the DOCS1 kinase domain. We have generated a new loss-of-function mutation in the DOCS1 gene (docs1-2) using the CRISPR-Cas9 technology. This new loss-of-function mutant and docs1-1 present similar phenotypes suggesting the original docs1-1 was a null allele. Besides the aluminum sensitivity phenotype, both docs1 mutants shared also several root phenotypes described previously: less root hairs and mixed identities of the outer cell layers. Moreover, our new results suggest that DOCS1 could also play a role in root cap development. We hypothesized these docs1 root phenotypes may affect gravity responses. As expected, in seedlings, the early gravitropic response was delayed. Furthermore, at adult stage, the root gravitropic set angle of docs1 mutants was also affected since docs1 mutant plants displayed larger root cone angles. All these observations add new insights into the DOCS1 gene function in gravitropic responses at several stages of plant development.
Bettembourg M, Dardou A, Audebert A, Thomas E, Frouin J, Guiderdoni E, Ahmadi N, Perin C, Dievart A, Courtois B (2017) Genome-wide association mapping for root cone angle in rice. Rice 10:45.
Dufayard JF, Bettembourg M, Fischer I, Droc G, Guiderdoni E, Périn C, Chantret N, Diévart A. (2017) New Insights on Leucine-Rich Repeats Receptor-Like Kinase Orthologous Relationships in Angiosperms. Front Plant Sci. 2017 Apr 5;8:381. Erratum in: Front Plant Sci. 2017 May 24;8:916.
Leucine-Rich Repeats Receptor-Like Kinase (LRR-RLK) genes represent a large and complex gene family in plants, mainly involved in development and stress responses. These receptors are composed of an LRR-containing extracellular domain (ECD), a transmembrane domain (TM) and an intracellular kinase domain (KD). To provide new perspectives on functional analyses of these genes in model and non-model plant species, we performed a phylogenetic analysis on 8,360 LRR-RLK receptors in 31 angiosperm genomes (8 monocots and 23 dicots). We identified 101 orthologous groups (OGs) of genes being conserved among almost all monocot and dicot species analyzed. We observed that more than 10% of these OGs are absent in the Brassicaceae species studied. We show that the ECD structural features are not always conserved among orthologs, suggesting that functions may have diverged in some OG sets. Moreover, we looked at targets of positive selection footprints in 12 pairs of OGs and noticed that depending on the subgroups, positive selection occurred more frequently either in the ECDs or in the KDs.
Henry S, Dievart A, Fanchon D, Pauluzzi G, Meynard D, Swarup R, Wu S, Lee CM, Gallagher K, Périn C. (2017) SHR overexpression induces the formation of supernumerary cell layers with cortex cell identity in rice. Dev Biol. 1;425(1):1-7.
Fischer I, Dievart A, Droc G, Dufayard JF, Chantret N. (2016) Evolutionary Dynamics of the Leucine-Rich Repeat Receptor-Like Kinase (LRR-RLK) Subfamily in Angiosperms. Plant Physiology 170 (3): 1595-610.
Gene duplications are an important factor in plant evolution, and lineage-specific expanded (LSE) genes are of particular interest. Receptor-like kinases expanded massively in land plants, and leucine-rich repeat receptor-like kinases (LRR-RLK) constitute the largest receptor-like kinases family. Based on the phylogeny of 7,554 LRR-RLK genes from 31 fully sequenced flowering plant genomes, the complex evolutionary dynamics of this family was characterized in depth. We studied the involvement of selection during the expansion of this family among angiosperms. LRR-RLK subgroups harbor extremely contrasting rates of duplication, retention, or loss, and LSE copies are predominantly found in subgroups involved in environmental interactions. Expansion rates also differ significantly depending on the time when rounds of expansion or loss occurred on the angiosperm phylogenetic tree. Finally, using a dN/dS-based test in a phylogenetic framework, we searched for selection footprints on LSE and single-copy LRR-RLK genes. Selective constraint appeared to be globally relaxed at LSE genes, and codons under positive selection were detected in 50% of them. Moreover, the leucine-rich repeat domains, and specifically four amino acids in them, were found to be the main targets of positive selection. Here, we provide an extensive overview of the expansion and evolution of this very large gene family.
Henry S, Divol F, Bettembourg M, Bureau C, Guiderdoni E, Perin C, Dievart A (2016) Immunoprofiling of Rice Root Cortex Reveals Two Cortical Subdomains. Frontiers in Plant Science 6:1139.
Dievart A., Perin C., Hirsch J., Bettembourg M., Lanau N., Artus F., Bureau C., Noel N., Droc G., Peyramard M., Pereira S., Courtois B., Morel J-B., Guiderdoni E. (2016) The phenome analysis of mutant alleles in Leucine-Rich Repeat Receptor-Like Kinase genes in rice reveals new potential targets for stress tolerant cereals. Plant science 242, 240-249.
We report here a systematic analysis of the role of the members of the LRR-RLK gene family by mutant phenotyping in the monocotyledon model plant Oryza sativa. We have targeted 176 of the ∼320 LRR-RLK genes (55.7%) and genotyped 288 mutant lines. Position of the insertion was confirmed in 128 lines corresponding to 100 LRR-RLK genes (31.6% of the entire family). All mutant lines harboring homozygous insertions have been screened for phenotypes under normal conditions and under various abiotic stresses. Mutant plants have been observed at several stages of growth, from seedlings in Petri dishes to flowering and grain filling under greenhouse conditions. Our results show that 37 of the LRR-RLK rice genes are potential targets for improvement especially in the generation of abiotic stress tolerant cereals.Lartaud M., Perin C., Courtois B., Thomas E., Henry S., Bettembourg M., Divol F., Lanau N., Artus F., Bureau C., Verdeil J.L., Sarah G., Guiderdoni E. and Dievart A. (2015) PHIV-RootCell: a supervised image analysis tool for rice root anatomical parameter quantification.Frontiers in Plant Science 5:790.
The PHIV-RootCell program is an imageJ toolset dedicated to the analysis of several root anatomical parameters based on images of rice root transverse sections. The program tracks root tissues and cell walls to measure areas and numbers of cells in cell files. PHIV-RootCell uses a semi-automated approach where the user has to supervise each step of the process and can proceed to corrections if not satisfied with the software's proposals. Data are exported as tabulated text files that can be directly used for statistical analyses.
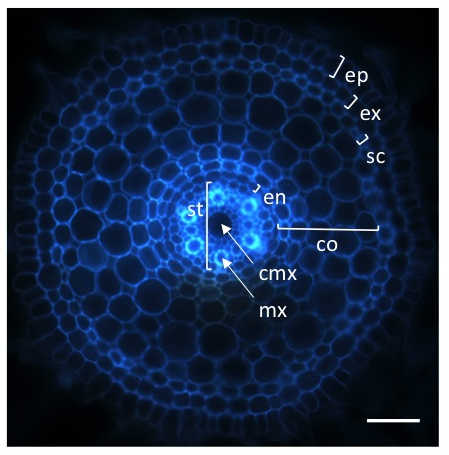
Autofluorescence image of a transverse histological root section. The rice root tissues are concentric cylinders; from external to internal, the cell layers are the epidermis (ep), exodermis (ex), sclerenchyma (sc), several layers of cortex (co), endodermis (en) and pericycle. The pericycle delimits the stele (st) containing the vascular vessels (the central metaxylem (cmx) and the metaxylems (mx)). Scale bar: 100 μm.

Screen capture of the polar transformed image from the PHIV-RootCell toolset. The center for the polar transformation is given by the stele selection. In this way, the metaxylem vessels are at the top of the image and the epidermis at the bottom.
Lorieux M, Blein M, Lozano J, Bouniol M, Droc G, Diévart A, Périn C, Mieulet D, Lanau N, Bès M, Rouvière C, Gay C, Piffanelli P, Larmande P, Michel C, Barnola I, Biderre-Petit C, Sallaud C, Perez P, Bourgis F, Ghesquière A, Gantet P, Tohme J, Morel JB, Guiderdoni E. (2012) In-depth molecular and phenotypic characterization in a rice insertion line library facilitates gene identification through reverse and forward genetics approaches. Plant Biotechnol Journal 10(5):555-68.
Diévart A, Gilbert N, Droc G, Attard A, Gourgues M, Guiderdoni E and Périn C. (2011) Leucine-Rich Repeats Receptor Kinases are sporadically distributed in eukaryotic genomes. BMC Evol Biol. 11(1):367.In this report, we show that LRR-RK subfamilies have been reinvented in several eukaryotic genomes outside plants.

Schematic phylogenetic representation of all of the genomes analyzed. The number of genomes in which LRR-RKs or LRR-RLKs (land plants) were found is followed in parentheses by the total number of genomes analyzed for each kingdom. On the right, the number of LRR-RLKs is given following the name of each species. Among the green algae, 6 genomes were studied; only one (Chlorella variabilis NC64A) contains LRR-RK proteins. Among the oomycetes, 1 genome of Saprolegniales (Saprolegnia parasitica) and 4 genomes of Peronosporales (Pythium ultimum, Phytophthora infestans, Phytophthora ramorum and Phytophthora sojae) were analyzed. Only proteins with an LRR-TM-KD organization are considered to be LRR-RKs. 1Soanes et al. 2010, 2Cock et al. 2010, 3this study, N.D. Not Determined.
Droc G, Ruiz M, Larmande P, Pereira A, Piffanelli P, Morel JB, Diévart A, Courtois B, Guiderdoni E and Perin C (2006) OryGenesDB: a database for rice reverse genetics. Nucleic Acid Research. 34: D736-40.
Diévart A, Hymes M, Li J, and Clark SE (2006) Brassinosteroid-independent function of BRI1/CLV1 chimeric receptors. Functional Plant Biology. 33 (8): 723-730.
Diévart A, Dalal M, Tax FE, Lacey AD, Huttly A, Li J, and Clark SE (2003) CLAVATA1 dominant-negative alleles reveal functional overlap between multiple receptor kinases regulating meristem and organ development. The Plant Cell. 15:1198-1211.
Hu C, Diévart A, Lupien M, Calvo E, Tremblay G and Jolicoeur P (2006) Overexpression of activated murine Notch1 and Notch3 in transgenic mice blocks mammary gland development and induces mammary tumors. American Journal of Pathology. 168(3): 973-990.
Lupien M, Diévart A, Morales CR, Hermo L, Calvo E, Kay DG, Hu C and Jolicoeur P (2006) Expression of constitutively active Notch1 in male genital tracts results in ectopic growth and blockage of efferent duct, junction, epididymal hyperplasia and sterility. Developmental Biology. 300 (2): 497-511.
Diévart A, Beaulieu N and Jolicoeur P (1999) Involvement of Notch1 in the development of mouse mammary tumors. Oncogene. 18:5973-5981.
Reviews
Ahmadi N., Audebert A., Bennett M.J., Bishopp A., Costa de Oliveira A., Courtois B., Diedhiou A., Diévart A., Gantet P., Ghesquière A., Guiderdoni E., Henry A., Inukai Y., Kochian L., Laplaze L., Lucas M., Luu D.T., Manneh B., Mo X., Muthurajan R., Périn C., Price A., Robin S., Sentenac H., Sine B., Uga Y., Véry A.A., Wissuwa M., Wu P., Xu J. (2014) The roots of future rice harvests. Rice 7:29.
Mieulet D, Diévart A, Droc G, Lanau N and Guiderdoni E. (2013) Reverse genetics in rice using Tos17. Book chapter for Methods in Molecular Biology: Plant Transposable Elements. 1057:205-21
Dievart A., Coudert Y., Gantet P., Pauluzzi F., Puig J., Divol F., Ahmadi N., Courtois B., Guiderdoni E., Perin C. (2013) Dissection des bases biologiques de caractères d'intérêt chez le riz : architecture et développement du système racinaire = Dissecting the biological bases of traits of interest in rice: Architecture and development of the root system. Cahiers agricultures 22 (5):475-483.
Meynard D., Mieulet D., Siré C., Petit J., Breitler J.C., Périn C., Diévart A., Divol F., Courtois B., Verdeil J.L., Gantet P., Guiderdoni E. (2013)La transformation génétique du riz: Méthodes, outil pour l'analyse fonctionnelle des genes et applications à l'amélioration variétale. Cahiers agricultures 22 (5):484-493.
Pauluzzi G, Divol F, Puig J, Guiderdoni E, Dievart A, Périn C. (2012) Surfing along the root ground tissue gene network. Developmental Biology May 1;365(1):14-22.
Rebouillat J, Dievart A, Verdeil J-L, Escoute J, Giese G, Breitler J-C, Gantet P, Espeout S, Guiderdoni E and Périn C (2009) Molecular Genetics of Rice Root Development. Rice. 2:15-34.
Diévart A and Clark SE (2004) LRR-containing receptors regulating plant development and defense. Development. 131 :251-261.
Diévart A and Clark SE (2003) Using mutant alleles to determine the structure and function of leucine-rich repeat receptor-like kinases. Current Opinion in Plant Biology, 6(5):507-16.
Jolicoeur P, Bouchard L, Guimond A, Ste-Marie M, Hanna Z and Diévart A (1998) Use of mouse mammary tumour virus (MMTV)/neu transgenic mice to identify genes collaborating with the c-erbB-2 oncogene in mammary tumour development. Biochem.Soc. Symp. 63, 159-165.Collaborations
Reshetnyak G, Jacobs JM, Auguy F, Sciallano C, Claude L, Medina C, Perez-Quintero AL, Comte A, Thomas E, Bogdanove A, Koebnik R, Szurek B, Dievart A, Brugidou C, Lacombe S, Cunnac S. (2021) An atypical class of non-coding small RNAs is produced in rice leaves upon bacterial infection. Sci Rep. 11(1):24141. doi: 10.1038/s41598-021-03391-9.
Zhang M,Kim Y, Zong J, Lin H, Dievart A, Li H, Zhang D, Liang W. (2019) Genome-wide analysis of the barley non-specific lipid transfer protein gene family. The Crop Journal 7(1): 65-76.
Plomion C, Aury JM, Amselem J, Leroy T, Murat F, Duplessis S, Faye S, Francillonne N, Labadie K, Le Provost G, Lesur I, Bartholomé J, Faivre-Rampant P, Kohler A, Leplé JC, Chantret N, Chen J, Diévart A, Alaeitabar T, Barbe V, Belser C, Bergès H, Bodénès C, Bogeat-Triboulot MB, Bouffaud ML, Brachi B, Chancerel E, Cohen D, Couloux A, Da Silva C, Dossat C, Ehrenmann F, Gaspin C, Grima-Pettenati J, Guichoux E, Hecker A, Herrmann S, Hugueney P, Hummel I, Klopp C, Lalanne C, Lascoux M, Lasserre E, Lemainque A, Desprez-Loustau ML, Luyten I, Madoui MA, Mangenot S, Marchal C, Maumus F, Mercier J, Michotey C, Panaud O, Picault N, Rouhier N, Rué O, Rustenholz C, Salin F, Soler M, Tarkka M, Velt A, Zanne AE, Martin F, Wincker P, Quesneville H, Kremer A, Salse J. (2018) Oak genome reveals facets of long lifespan. Nat Plants. 4(7):440-452.
Giri J, Bhosale R, Huang G, Pandey BK, Parker H, Zappala S, Yang J, Dievart A, Bureau C, Ljung K, Price A, Rose T, Larrieu A, Mairhofer S, Sturrock CJ, White P, Dupuy L, Hawkesford M, Perin C, Liang W, Peret B, Hodgman CT, Lynch J, Wissuwa M, Zhang D, Pridmore T, Mooney SJ, Guiderdoni E, Swarup R and Bennett MJ (2018) Rice auxin influx carrier OsAUX1 facilitates root hair elongation in response to low external phosphate. Nat comm. 12;9(1):1408.
Tsai WC, Dievart A, Hsu CC, Hsiao YY, Chiou SY,
Huang H, Chen HH (2017) Post genomics
era for orchid research. Botanical Studies. 58:61.
Delteil A, Gobbato E, Cayrol B, Estevan J, Michel-Romiti C, Dievart A, Kroj T, Morel JB. (2016) Several wall-associated kinases participate positively and negatively in basal defense against rice blast fungus. BMC Plant Biol. 16:17.
Cai J, Liu X, Vanneste K, Proost S, Tsai WC, Liu KW, Chen LJ, He Y, Xu Q, Bian C, Zheng Z, Sun F, Liu W, Hsiao YY, Pan ZJ, Hsu CC, Yang YP, Hsu YC, Chuang YC, Dievart A, Dufayard JF, Xu X, Wang JY, Wang J, Xiao XJ, Zhao XM, Du R, Zhang GQ, Wang M, Su YY, Xie GC, Liu GH, Li LQ, Huang LQ, Luo YB, Chen HH, Van de Peer Y, Liu ZJ. (2015) The genome sequence of the orchid Phalaenopsis equestris. Nature Genetics47(1):65-72. doi: 10.1038/ng.3149.
Coudert Y, Dievart A, Droc G, Gantet P. (2013) ASL/LBD Phylogeny Suggests that Genetic Mechanisms of Root Initiation Downstream of Auxin Are Distinct in Lycophytes and Euphyllophytes. Molecular Biology and Evolution 30(3):569-72.
D'Hont A, Denoeud F, Aury JM, Baurens FC, Carreel F, Garsmeur O, Noel B, Bocs S, Droc G, Rouard M, Da Silva C, Jabbari K, Cardi C, Poulain J, Souquet M, Labadie K, Jourda C, Lengellé J, Rodier-Goud M, Alberti A, Bernard M, Correa M, Ayyampalayam S, Mckain MR, Leebens-Mack J, Burgess D, Freeling M, Mbéguié-A-Mbéguié D, Chabannes M, Wicker T, Panaud O, Barbosa J, Hribova E, Heslop-Harrison P, Habas R, Rivallan R, Francois P, Poiron C, Kilian A, Burthia D, Jenny C, Bakry F, Brown S, Guignon V, Kema G, Dita M, Waalwijk C, Joseph S, Dievart A, Jaillon O, Leclercq J, Argout X, Lyons E, Almeida A, Jeridi M, Dolezel J, Roux N, Risterucci AM, Weissenbach J, Ruiz M, Glaszmann JC, Quétier F, Yahiaoui N, Wincker P. (2012) The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature. 488(7410):213-7.
Argout X, Salse J, Aury JM, Guiltinan MJ, Droc G, Gouzy J, Allegre M, Chaparro C, Legavre T, Maximova SN, Abrouk M, Murat F, Fouet O, Poulain J, Ruiz M, Roguet Y, Rodier-Goud M, Barbosa-Neto JF, Sabot F, Kudrna D, Ammiraju JS, Schuster SC, Carlson JE, Sallet E, Schiex T, Dievart A, Kramer M, Gelley L, Shi Z, Bérard A, Viot C, Boccara M, Risterucci AM, Guignon V, Sabau X, Axtell MJ, Ma Z, Zhang Y, Brown S, Bourge M, Golser W, Song X, Clement D, Rivallan R, Tahi M, Akaza JM, Pitollat B, Gramacho K, D'Hont A, Brunel D, Infante D, Kebe I, Costet P, Wing R, McCombie WR, Guiderdoni E, Quetier F, Panaud O, Wincker P, Bocs S, Lanaud C. (2011) The genome of Theobroma cacao. Nat Genet. 43(2):101-8. Epub 2010 Dec 26.
Anukul N, Ramos RA, Mehrshahi P, Castelazo AS, Parker H, Dievart A, Lanau N, Mieulet D, Tucker G, Guideroni E., Barrett D.A. and Bennett M.J. (2010) Folate Polyglutamylation is Required for Rice Seed Development. Rice, 13:1939-8425.